
News and Announcements from the CDB
The most striking feature of the mammalian brain is the presence of a large, complex cerebral cortex, a six-layered structure comprising different neuronal types and each layer being derived from self-renewing neuroepithelial cells, called apical progenitors (APs), found in the ventricular zone. APs initially proliferate through symmetric cell division, but later switch to differentiation mode, undergoing asymmetric cell division to produce one AP that will continue to self-renew and another daughter cell that becomes an intermediate progenitor (IP) which will differentiate to give rise to neurons. As the type of neurons born from IPs changes according to progression of time, from deep-layer neurons to more upper-layer neurons, the precursor APs are thought to “sense” temporal progression of development to change fate of their differentiating progeny, but the underlying mechanism regulating time in the cell remains unknown.
Now a collaborative study between research scientist Daijiro Konno of the Laboratory for Cell Asymmetry (Fumio Matsuzaki, Team Leader), Ayano Kawaguchi, an alumnus of the laboratory and currently associate professor at Nagoya University, and others has identified a group of so-called “temporal-axis genes” in neural progenitors—genes that show fluctuations in expression levels only with respect to progression of developmental time—using the mouse model. They used single cell transcriptomics to analyze changes in gene expression at several different developmental stages, and also revealed that temporal expression patterns of the temporal-axis genes were unaffected by cell-cycle arrest, thus functioning independent to cell cycle. Their findings were published in the online journal, Nature Communications.
Identifying the molecular factors that regulate the timing when APs temporally change their properties, including switch from proliferation to neuronal production, has been a challenge as this process is not synchronized. Thus it is difficult to make a distinction between genes involved in temporal change of degree of differentiation and those involved only in shift of temporal pattern of AP identity. In this study, the research group decided to apply single cell transcriptomics to comprehensively analyze and compare all genes transcribed within a cell at several different developmental stages (E11, E14 and E16). From their transcriptome data, they used a statistical method called principal component analysis (PCA) to identify a set of genes that showed changes in gene expression only in correlation to temporal progression. These changes in gene expression patterns were independent to differential fate of progenitors (whether the cell is an AP or an IP), and most of the temporal-axis genes displayed the largest changes in expression around E12.
The cell cycle would appear as the most likely mechanism involved in regulating time within a cell since it is a fundamental mechanism found in all cells, with genes being activated in turn to prompt cells to steadily advance to the next stage of the cell cycle. The group devised an experimental system to transiently arrest the cell cycle of APs in the embryonic mouse brain by activating and terminating expression of genes involved in cell-cycle progression via electroporation, and then tracked the effects of transient cell-cycle arrest on expression patterns of temporal-axis genes. They found that, despite the temporary cell-cycle arrest, expression patterns of the temporal-axis genes, which displayed considerable changes in expression around E12, remained unchanged. Furthermore, when neuronal types that are born after transient cell-cycle arrest were examined, the team noticed that these APs failed to produce deep-layer neurons, and instead differentiated into the same type of upper-layer neurons that would be produced under normal conditions if cell-cycle had not been arrested. Together, these findings suggest that the ability of APs to produce sequentially diverse neurons is independent to the cell cycle.
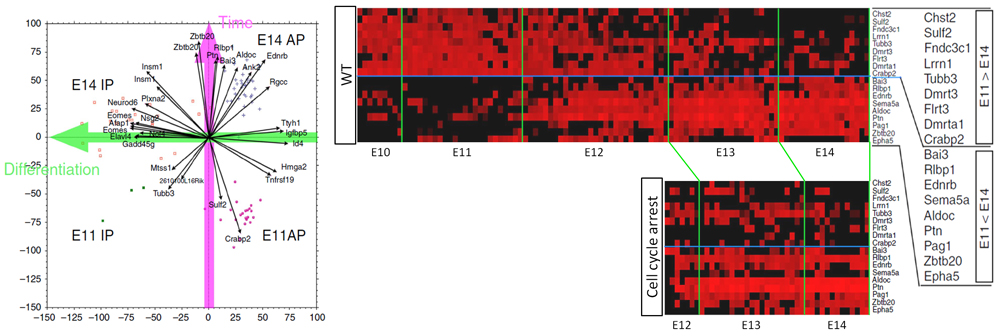
“Our experiments also suggested the existence of two types of temporal-axis genes, those that can be controlled cell intrinsically and those that require an extrinsic signal to trigger gene expression changes,” explains Matsuzaki. “Whereas the question of how living organisms keep time has remained largely a mystery, our present study presents evidence refuting the widely-held hypothesis of the involvement of the cell-cycle, at least in the context of brain development. Our next step is to identify the key temporal-axis genes, and eventually reveal the complete mechanism controlling temporal progression in cells.”
| Link to article |
Cell-cycle-independent transitions in temporal identity of mammalian neural progenitor cells. |
|---|